04. 2D CFAR
2D CFAR
- Determine the number of Training cells for each dimension. Similarly, pick the number of guard cells.
- Slide the cell under test across the complete matrix. Make sure the CUT has margin for Training and Guard cells from the edges.
- For every iteration sum the signal level within all the training cells. To sum convert the value from logarithmic to linear using db2pow function.
- Average the summed values for all of the training cells used. After averaging convert it back to logarithmic using pow2db.
- Further add the offset to it to determine the threshold.
- Next, compare the signal under CUT against this threshold.
- If the CUT level > threshold assign it a value of 1, else equate it to 0.
The process above will generate a thresholded block, which is smaller than the Range Doppler Map as the CUTs cannot be located at the edges of the matrix due to the presence of Target and Guard cells. Hence, those cells will not be thresholded.
- To keep the map size same as it was before CFAR, equate all the non-thresholded cells to 0.
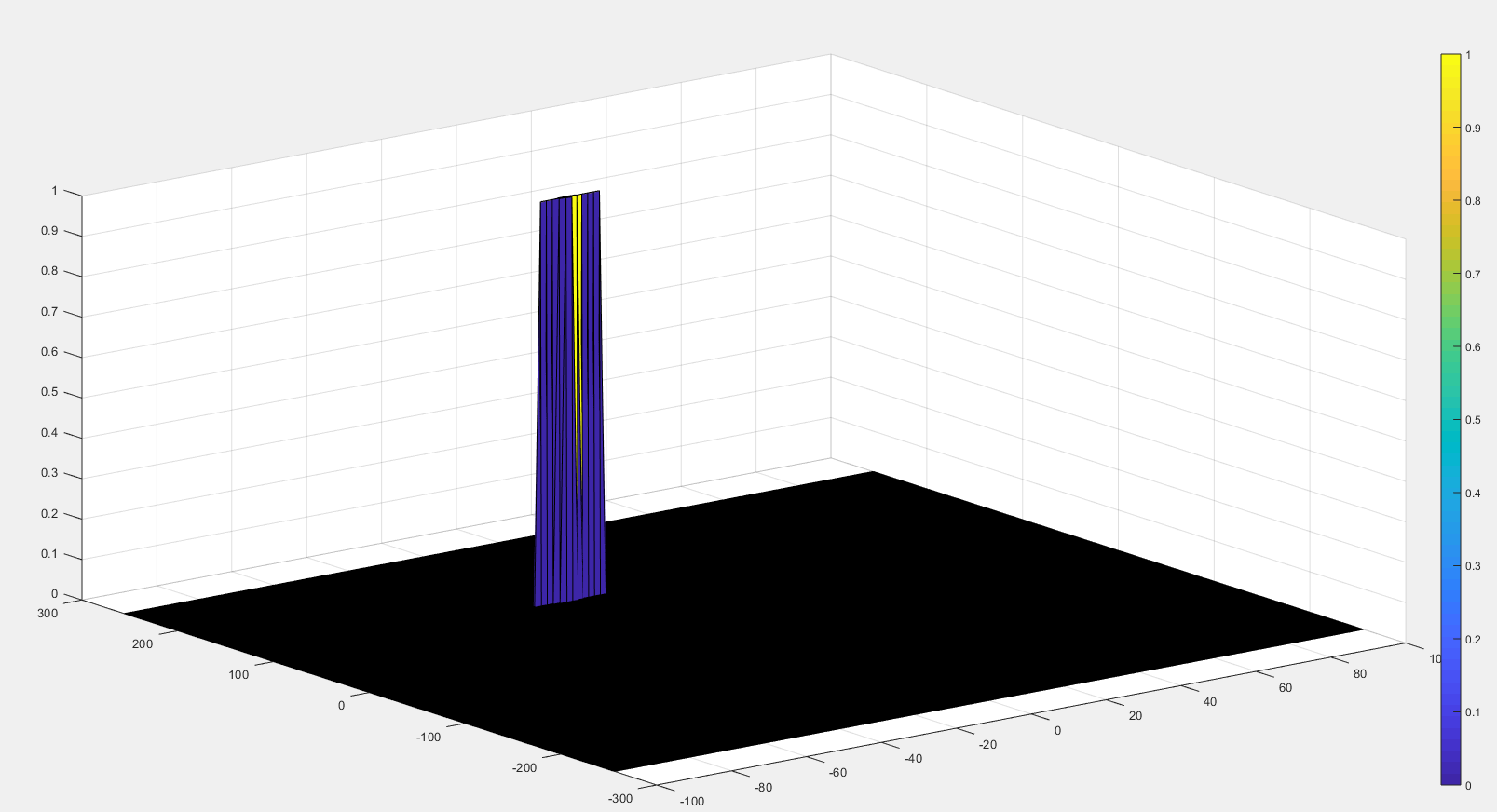
The output of the 2D CFAR process
Once you have completed this, you are done. Congratulations on doing a great job on this final project!